
Like most people in the world right now, I’m genuinely concerned about COVID-19. I find myself constantly analyzing my personal health and wondering if/when I will contract it.
The more I worry about it, the more it turns into a painful mind game of legitimate symptoms combined with hypochondria:
- I woke up this morning feeling a bit achy and run down.
- As I pulled myself out of bed, I noticed my nose was running (although it’s now reported that a runny nose is not a symptom of COVID-19).
- By the time I made it to the bathroom to grab a tissue, I was coughing as well.
At first, I didn’t think much of it — I have pollen allergies and due to the warm weather on the eastern coast of the United States, spring has come early this year. My allergies were likely just acting up.
But my symptoms didn’t improve throughout the day.
I’m actually sitting here, writing the this tutorial, with a thermometer in my mouth; and glancing down I see that it reads 99.4° Fahrenheit.
My body runs a bit cooler than most, typically in the 97.4°F range. Anything above 99°F is a low-grade fever for me.
Cough and low-grade fever? That could be COVID-19…or it could simply be my allergies.
It’s impossible to know without a test, and that “not knowing” is what makes this situation so scary from a visceral human level.
As humans, there is nothing more terrifying than the unknown.
Despite my anxieties, I try to rationalize them away. I’m in my early 30s, very much in shape, and my immune system is strong. I’ll quarantine myself (just in case), rest up, and pull through just fine — COVID-19 doesn’t scare me from my own personal health perspective (at least that’s what I keep telling myself).
That said, I am worried about my older relatives, including anyone that has pre-existing conditions, or those in a nursing home or hospital. They are vulnerable and it would be truly devastating to see them go due to COVID-19.
Instead of sitting idly by and letting whatever is ailing me keep me down (be it allergies, COVID-19, or my own personal anxieties), I decided to do what I do best — focus on the overall CV/DL community by writing code, running experiments, and educating others on how to use computer vision and deep learning in practical, real-world applications.
That said, I’ll be honest, this is not the most scientific article I’ve ever written. Far from it, in fact. The methods and datasets used would not be worthy of publication. But they serve as a starting point for those who need to feel like they’re doing something to help.
I care about you and I care about this community. I want to do what I can to help — this blog post is my way of mentally handling a tough time, while simultaneously helping others in a similar situation.
I hope you see it as such.
Inside of today’s tutorial, you will learn how to:
- Sample an open source dataset of X-ray images for patients who have tested positive for COVID-19
- Sample “normal” (i.e., not infected) X-ray images from healthy patients
- Train a CNN to automatically detect COVID-19 in X-ray images via the dataset we created
- Evaluate the results from an educational perspective
Disclaimer: I’ve hinted at this already but I’ll say it explicitly here. The methods and techniques used in this post are meant for educational purposes only. This is not a scientifically rigorous study, nor will it be published in a journal. This article is for readers who are interested in (1) Computer Vision/Deep Learning and want to learn via practical, hands-on methods and (2) are inspired by current events. I kindly ask that you treat it as such.
To learn how you could detect COVID-19 in X-ray images by using Keras, TensorFlow, and Deep Learning, just keep reading!
Detecting COVID-19 in X-ray images with Keras, TensorFlow, and Deep Learning
In the first part of this tutorial, we’ll discuss how COVID-19 could be detected in chest X-rays of patients.
From there, we’ll review our COVID-19 chest X-ray dataset.
I’ll then show you how to train a deep learning model using Keras and TensorFlow to predict COVID-19 in our image dataset.
Disclaimer
This blog post on automatic COVID-19 detection is for educational purposes only. It is not meant to be a reliable, highly accurate COVID-19 diagnosis system, nor has it been professionally or academically vetted.
My goal is simply to inspire you and open your eyes to how studying computer vision/deep learning and then applying that knowledge to the medical field can make a big impact on the world.
Simply put: You don’t need a degree in medicine to make an impact in the medical field — deep learning practitioners working closely with doctors and medical professionals can solve complex problems, save lives, and make the world a better place.
My hope is that this tutorial inspires you to do just that.
But with that said, researchers, journal curators, and peer review systems are being overwhelmed with submissions containing COVID-19 prediction models of questionable quality. Please do not take the code/model from this post and submit it to a journal or Open Science — you’ll only add to the noise.
Furthermore, if you intend on performing research using this post (or any other COVID-19 article you find online), make sure you refer to the TRIPOD guidelines on reporting predictive models.
As you’re likely aware, artificial intelligence applied to the medical domain can have very real consequences. Only publish or deploy such models if you are a medical expert, or closely consulting with one.
How could COVID-19 be detected in X-ray images?
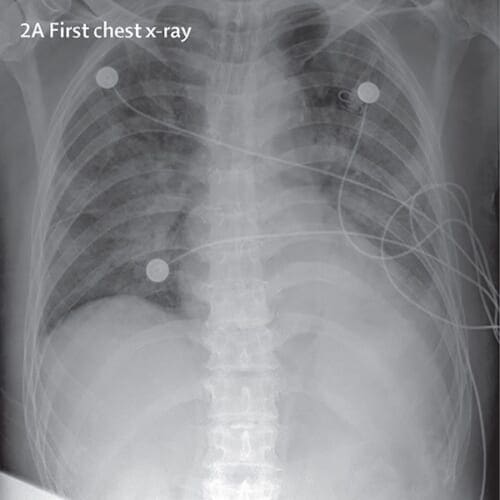
COVID-19 tests are currently hard to come by — there are simply not enough of them and they cannot be manufactured fast enough, which is causing panic.
When there’s panic, there are nefarious people looking to take advantage of others, namely by selling fake COVID-19 test kits after finding victims on social media platforms and chat applications.
Given that there are limited COVID-19 testing kits, we need to rely on other diagnosis measures.
For the purposes of this tutorial, I thought to explore X-ray images as doctors frequently use X-rays and CT scans to diagnose pneumonia, lung inflammation, abscesses, and/or enlarged lymph nodes.
Since COVID-19 attacks the epithelial cells that line our respiratory tract, we can use X-rays to analyze the health of a patient’s lungs.
And given that nearly all hospitals have X-ray imaging machines, it could be possible to use X-rays to test for COVID-19 without the dedicated test kits.
A drawback is that X-ray analysis requires a radiology expert and takes significant time — which is precious when people are sick around the world. Therefore developing an automated analysis system is required to save medical professionals valuable time.
Note: There are newer publications that suggest CT scans are better for diagnosing COVID-19, but all we have to work with for this tutorial is an X-ray image dataset. Secondly, I am not a medical expert and I presume there are other, more reliable, methods that doctors and medical professionals will use to detect COVID-19 outside of the dedicated test kits.
Our COVID-19 patient X-ray image dataset
The COVID-19 X-ray image dataset we’ll be using for this tutorial was curated by Dr. Joseph Cohen, a postdoctoral fellow at the University of Montreal.
One week ago, Dr. Cohen started collecting X-ray images of COVID-19 cases and publishing them in the following GitHub repo.
Inside the repo you’ll find example of COVID-19 cases, as well as MERS, SARS, and ARDS.
In order to create the COVID-19 X-ray image dataset for this tutorial, I:
- Parsed the
metadata.csvfile found in Dr. Cohen’s repository. - Selected all rows that are:
- Positive for COVID-19 (i.e., ignoring MERS, SARS, and ARDS cases).
- Posterioranterior (PA) view of the lungs. I used the PA view as, to my knowledge, that was the view used for my “healthy” cases, as discussed below; however, I’m sure that a medical professional will be able clarify and correct me if I am incorrect (which I very well may be, this is just an example).
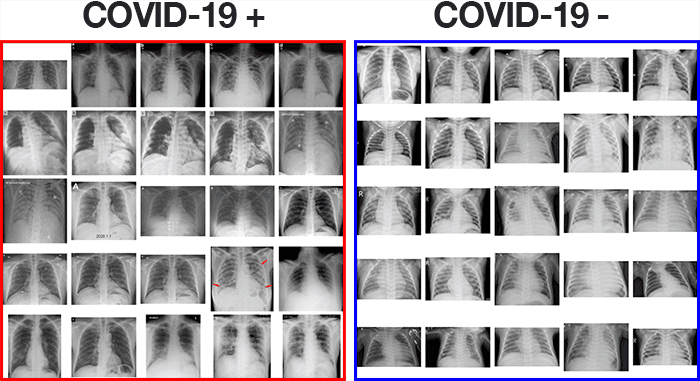
In total, that left me with 25 X-ray images of positive COVID-19 cases (Figure 2, left).
The next step was to sample X-ray images of healthy patients.
To do so, I used Kaggle’s Chest X-Ray Images (Pneumonia) dataset and sampled 25 X-ray images from healthy patients (Figure 2, right). There are a number of problems with Kaggle’s Chest X-Ray dataset, namely noisy/incorrect labels, but it served as a good enough starting point for this proof of concept COVID-19 detector.
After gathering my dataset, I was left with 50 total images, equally split with 25 images of COVID-19 positive X-rays and 25 images of healthy patient X-rays.
I’ve included my sample dataset in the “Downloads” section of this tutorial, so you do not have to recreate it.
Additionally, I have included my Python scripts used to generate the dataset in the downloads as well, but these scripts will not be reviewed in this tutorial as they are outside the scope of the post.
Project structure
Go ahead and grab today’s code and data from the “Downloads” section of this tutorial. From there, extract the files and you’ll be presented with the following directory structure:
$ tree --dirsfirst --filelimit 10 . ├── dataset │ ├── covid [25 entries] │ └── normal [25 entries] ├── build_covid_dataset.py ├── sample_kaggle_dataset.py ├── train_covid19.py ├── plot.png └── covid19.model 3 directories, 5 files
Our coronavirus (COVID-19) chest X-ray data is in the dataset/ directory where our two classes of data are separated into covid/ and normal/.
Both of my dataset building scripts are provided; however, we will not be reviewing them today.
Instead, we will review the train_covid19.py script which trains our COVID-19 detector.
Let’s dive in and get to work!
Implementing our COVID-19 training script using Keras and TensorFlow
Now that we’ve reviewed our image dataset along with the corresponding directory structure for our project, let’s move on to fine-tuning a Convolutional Neural Network to automatically diagnose COVID-19 using Keras, TensorFlow, and deep learning.
Open up the train_covid19.py file in your directory structure and insert the following code:
# import the necessary packages from tensorflow.keras.preprocessing.image import ImageDataGenerator from tensorflow.keras.applications import VGG16 from tensorflow.keras.layers import AveragePooling2D from tensorflow.keras.layers import Dropout from tensorflow.keras.layers import Flatten from tensorflow.keras.layers import Dense from tensorflow.keras.layers import Input from tensorflow.keras.models import Model from tensorflow.keras.optimizers import Adam from tensorflow.keras.utils import to_categorical from sklearn.preprocessing import LabelBinarizer from sklearn.model_selection import train_test_split from sklearn.metrics import classification_report from sklearn.metrics import confusion_matrix from imutils import paths import matplotlib.pyplot as plt import numpy as np import argparse import cv2 import os
This script takes advantage of TensorFlow 2.0 and Keras deep learning libraries via a selection of tensorflow.keras imports.
Additionally, we use scikit-learn, the de facto Python library for machine learning, matplotlib for plotting, and OpenCV for loading and preprocessing images in the dataset.
To learn how to install TensorFlow 2.0 (including relevant scikit-learn, OpenCV, and matplotlib libraries), just follow my Ubuntu or macOS guide.
With our imports taken care of, next we will parse command line arguments and initialize hyperparameters:
# construct the argument parser and parse the arguments
ap = argparse.ArgumentParser()
ap.add_argument("-d", "--dataset", required=True,
help="path to input dataset")
ap.add_argument("-p", "--plot", type=str, default="plot.png",
help="path to output loss/accuracy plot")
ap.add_argument("-m", "--model", type=str, default="covid19.model",
help="path to output loss/accuracy plot")
args = vars(ap.parse_args())
# initialize the initial learning rate, number of epochs to train for,
# and batch size
INIT_LR = 1e-3
EPOCHS = 25
BS = 8
Our three command line arguments (Lines 24-31) include:
--dataset: The path to our input dataset of chest X-ray images.--plot: An optional path to an output training history plot. By default the plot is namedplot.pngunless otherwise specified via the command line.--model: The optional path to our output COVID-19 model; by default it will be namedcovid19.model.
From there we initialize our initial learning rate, number of training epochs, and batch size hyperparameters (Lines 35-37).
We’re now ready to load and preprocess our X-ray data:
# grab the list of images in our dataset directory, then initialize
# the list of data (i.e., images) and class images
print("[INFO] loading images...")
imagePaths = list(paths.list_images(args["dataset"]))
data = []
labels = []
# loop over the image paths
for imagePath in imagePaths:
# extract the class label from the filename
label = imagePath.split(os.path.sep)[-2]
# load the image, swap color channels, and resize it to be a fixed
# 224x224 pixels while ignoring aspect ratio
image = cv2.imread(imagePath)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (224, 224))
# update the data and labels lists, respectively
data.append(image)
labels.append(label)
# convert the data and labels to NumPy arrays while scaling the pixel
# intensities to the range [0, 1]
data = np.array(data) / 255.0
labels = np.array(labels)
To load our data, we grab all paths to images in in the --dataset directory (Lines 42). Then, for each imagePath, we:
- Extract the class
label(eithercovidornormal) from the path (Line 49). - Load the
image, and preprocess it by converting to RGB channel ordering, and resizing it to 224×224 pixels so that it is ready for our Convolutional Neural Network (Lines 53-55). - Update our
dataandlabelslists respectively (Lines 58 and 59).
We then scale pixel intensities to the range [0, 1] and convert both our data and labels to NumPy array format (Lines 63 and 64).
Next we will one-hot encode our labels and create our training/testing splits:
# perform one-hot encoding on the labels lb = LabelBinarizer() labels = lb.fit_transform(labels) labels = to_categorical(labels) # partition the data into training and testing splits using 80% of # the data for training and the remaining 20% for testing (trainX, testX, trainY, testY) = train_test_split(data, labels, test_size=0.20, stratify=labels, random_state=42) # initialize the training data augmentation object trainAug = ImageDataGenerator( rotation_range=15, fill_mode="nearest")
One-hot encoding of labels takes place on Lines 67-69 meaning that our data will be in the following format:
[[0. 1.] [0. 1.] [0. 1.] ... [1. 0.] [1. 0.] [1. 0.]]
Each encoded label consists of a two element array with one of the elements being “hot” (i.e., 1) versus “not” (i.e., 0).
Lines 73 and 74 then construct our data split, reserving 80% of the data for training and 20% for testing.
In order to ensure that our model generalizes, we perform data augmentation by setting the random image rotation setting to 15 degrees clockwise or counterclockwise.
Lines 77-79 initialize the data augmentation generator object.
From here we will initialize our VGGNet model and set it up for fine-tuning:
# load the VGG16 network, ensuring the head FC layer sets are left # off baseModel = VGG16(weights="imagenet", include_top=False, input_tensor=Input(shape=(224, 224, 3))) # construct the head of the model that will be placed on top of the # the base model headModel = baseModel.output headModel = AveragePooling2D(pool_size=(4, 4))(headModel) headModel = Flatten(name="flatten")(headModel) headModel = Dense(64, activation="relu")(headModel) headModel = Dropout(0.5)(headModel) headModel = Dense(2, activation="softmax")(headModel) # place the head FC model on top of the base model (this will become # the actual model we will train) model = Model(inputs=baseModel.input, outputs=headModel) # loop over all layers in the base model and freeze them so they will # *not* be updated during the first training process for layer in baseModel.layers: layer.trainable = False
Lines 83 and 84 instantiate the VGG16 network with weights pre-trained on ImageNet, leaving off the FC layer head.
From there, we construct a new fully-connected layer head consisting of POOL => FC = SOFTMAX layers (Lines 88-93) and append it on top of VGG16 (Line 97).
We then freeze the CONV weights of VGG16 such that only the FC layer head will be trained (Lines 101-102); this completes our fine-tuning setup.
We’re now ready to compile and train our COVID-19 (coronavirus) deep learning model:
# compile our model
print("[INFO] compiling model...")
opt = Adam(lr=INIT_LR, decay=INIT_LR / EPOCHS)
model.compile(loss="binary_crossentropy", optimizer=opt,
metrics=["accuracy"])
# train the head of the network
print("[INFO] training head...")
H = model.fit_generator(
trainAug.flow(trainX, trainY, batch_size=BS),
steps_per_epoch=len(trainX) // BS,
validation_data=(testX, testY),
validation_steps=len(testX) // BS,
epochs=EPOCHS)
Lines 106-108 compile the network with learning rate decay and the Adam optimizer. Given that this is a 2-class problem, we use "binary_crossentropy" loss rather than categorical crossentropy.
To kick off our COVID-19 neural network training process, we make a call to Keras’ fit_generator method, while passing in our chest X-ray data via our data augmentation object (Lines 112-117).
Next, we’ll evaluate our model:
# make predictions on the testing set
print("[INFO] evaluating network...")
predIdxs = model.predict(testX, batch_size=BS)
# for each image in the testing set we need to find the index of the
# label with corresponding largest predicted probability
predIdxs = np.argmax(predIdxs, axis=1)
# show a nicely formatted classification report
print(classification_report(testY.argmax(axis=1), predIdxs,
target_names=lb.classes_))
For evaluation, we first make predictions on the testing set and grab the prediction indices (Lines 121-125).
We then generate and print out a classification report using scikit-learn’s helper utility (Lines 128 and 129).
Next we’ll compute a confusion matrix for further statistical evaluation:
# compute the confusion matrix and and use it to derive the raw
# accuracy, sensitivity, and specificity
cm = confusion_matrix(testY.argmax(axis=1), predIdxs)
total = sum(sum(cm))
acc = (cm[0, 0] + cm[1, 1]) / total
sensitivity = cm[0, 0] / (cm[0, 0] + cm[0, 1])
specificity = cm[1, 1] / (cm[1, 0] + cm[1, 1])
# show the confusion matrix, accuracy, sensitivity, and specificity
print(cm)
print("acc: {:.4f}".format(acc))
print("sensitivity: {:.4f}".format(sensitivity))
print("specificity: {:.4f}".format(specificity))
Here we:
- Generate a confusion matrix (Line 133)
- Use the confusion matrix to derive the accuracy, sensitivity, and specificity (Lines 135-137) and print each of these metrics (Lines 141-143)
We then plot our training accuracy/loss history for inspection, outputting the plot to an image file:
# plot the training loss and accuracy
N = EPOCHS
plt.style.use("ggplot")
plt.figure()
plt.plot(np.arange(0, N), H.history["loss"], label="train_loss")
plt.plot(np.arange(0, N), H.history["val_loss"], label="val_loss")
plt.plot(np.arange(0, N), H.history["accuracy"], label="train_acc")
plt.plot(np.arange(0, N), H.history["val_accuracy"], label="val_acc")
plt.title("Training Loss and Accuracy on COVID-19 Dataset")
plt.xlabel("Epoch #")
plt.ylabel("Loss/Accuracy")
plt.legend(loc="lower left")
plt.savefig(args["plot"])
Finally we serialize our tf.keras COVID-19 classifier model to disk:
# serialize the model to disk
print("[INFO] saving COVID-19 detector model...")
model.save(args["model"], save_format="h5")
Training our COVID-19 detector with Keras and TensorFlow
With our train_covid19.py script implemented, we are now ready to train our automatic COVID-19 detector.
Make sure you use the “Downloads” section of this tutorial to download the source code, COVID-19 X-ray dataset, and pre-trained model.
From there, open up a terminal and execute the following command to train the COVID-19 detector:
$ python train_covid19.py --dataset dataset
[INFO] loading images...
[INFO] compiling model...
[INFO] training head...
Epoch 1/25
5/5 [==============================] - 20s 4s/step - loss: 0.7169 - accuracy: 0.6000 - val_loss: 0.6590 - val_accuracy: 0.5000
Epoch 2/25
5/5 [==============================] - 0s 86ms/step - loss: 0.8088 - accuracy: 0.4250 - val_loss: 0.6112 - val_accuracy: 0.9000
Epoch 3/25
5/5 [==============================] - 0s 99ms/step - loss: 0.6809 - accuracy: 0.5500 - val_loss: 0.6054 - val_accuracy: 0.5000
Epoch 4/25
5/5 [==============================] - 1s 100ms/step - loss: 0.6723 - accuracy: 0.6000 - val_loss: 0.5771 - val_accuracy: 0.6000
...
Epoch 22/25
5/5 [==============================] - 0s 99ms/step - loss: 0.3271 - accuracy: 0.9250 - val_loss: 0.2902 - val_accuracy: 0.9000
Epoch 23/25
5/5 [==============================] - 0s 99ms/step - loss: 0.3634 - accuracy: 0.9250 - val_loss: 0.2690 - val_accuracy: 0.9000
Epoch 24/25
5/5 [==============================] - 27s 5s/step - loss: 0.3175 - accuracy: 0.9250 - val_loss: 0.2395 - val_accuracy: 0.9000
Epoch 25/25
5/5 [==============================] - 1s 101ms/step - loss: 0.3655 - accuracy: 0.8250 - val_loss: 0.2522 - val_accuracy: 0.9000
[INFO] evaluating network...
precision recall f1-score support
covid 0.83 1.00 0.91 5
normal 1.00 0.80 0.89 5
accuracy 0.90 10
macro avg 0.92 0.90 0.90 10
weighted avg 0.92 0.90 0.90 10
[[5 0]
[1 4]]
acc: 0.9000
sensitivity: 1.0000
specificity: 0.8000
[INFO] saving COVID-19 detector model...
Automatic COVID-19 diagnosis from X-ray image results
Disclaimer: The following section does not claim, nor does it intend to “solve”, COVID-19 detection. It is written in the context, and from the results, of this tutorial only. It is an example for budding computer vision and deep learning practitioners so they can learn about various metrics, including raw accuracy, sensitivity, and specificity (and the tradeoffs we must consider when working with medical applications). Again, this section/tutorial does not claim to solve COVID-19 detection.
As you can see from the results above, our automatic COVID-19 detector is obtaining ~90-92% accuracy on our sample dataset based solely on X-ray images — no other data, including geographical location, population density, etc. was used to train this model.
We are also obtaining 100% sensitivity and 80% specificity implying that:
- Of patients that do have COVID-19 (i.e., true positives), we could accurately identify them as “COVID-19 positive” 100% of the time using our model.
- Of patients that do not have COVID-19 (i.e., true negatives), we could accurately identify them as “COVID-19 negative” only 80% of the time using our model.
As our training history plot shows, our network is not overfitting, despite having very limited training data:
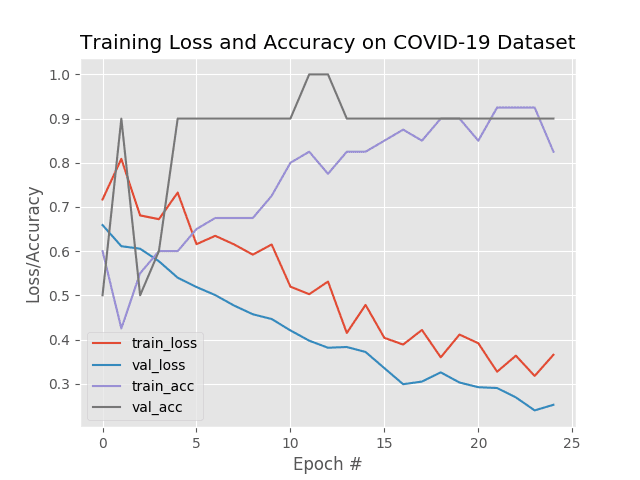
Being able to accurately detect COVID-19 with 100% accuracy is great; however, our true negative rate is a bit concerning — we don’t want to classify someone as “COVID-19 negative” when they are “COVID-19 positive”.
In fact, the last thing we want to do is tell a patient they are COVID-19 negative, and then have them go home and infect their family and friends; thereby transmitting the disease further.
We also want to be really careful with our false positive rate — we don’t want to mistakenly classify someone as “COVID-19 positive”, quarantine them with other COVID-19 positive patients, and then infect a person who never actually had the virus.
Balancing sensitivity and specificity is incredibly challenging when it comes to medical applications, especially infectious diseases that can be rapidly transmitted, such as COVID-19.
When it comes to medical computer vision and deep learning, we must always be mindful of the fact that our predictive models can have very real consequences — a missed diagnosis can cost lives.
Again, these results are gathered for educational purposes only. This article and accompanying results are not intended to be a journal article nor does it conform to the TRIPOD guidelines on reporting predictive models. I would suggest you refer to these guidelines for more information, if you are so interested.
Limitations, improvements, and future work

One of the biggest limitations of the method discussed in this tutorial is data.
We simply don’t have enough (reliable) data to train a COVID-19 detector.
Hospitals are already overwhelmed with the number of COVID-19 cases, and given patients rights and confidentiality, it becomes even harder to assemble quality medical image datasets in a timely fashion.
I imagine in the next 12-18 months we’ll have more high quality COVID-19 image datasets; but for the time being, we can only make do with what we have.
I have done my best (given my current mental state and physical health) to put together a tutorial for my readers who are interested in applying computer vision and deep learning to the COVID-19 pandemic given my limited time and resources; however, I must remind you that I am not a trained medical expert.
For the COVID-19 detector to be deployed in the field, it would have to go through rigorous testing by trained medical professionals, working hand-in-hand with expert deep learning practitioners. The method covered here today is certainly not such a method, and is meant for educational purposes only.
Furthermore, we need to be concerned with what the model is actually “learning”.
As I discussed in last week’s Grad-CAM tutorial, it’s possible that our model is learning patterns that are not relevant to COVID-19, and instead are just variations between the two data splits (i.e., positive versus negative COVID-19 diagnosis).
It would take a trained medical professional and rigorous testing to validate the results coming out of our COVID-19 detector.
And finally, future (and better) COVID-19 detectors will be multi-modal.
Right now we are using only image data (i.e., X-rays) — better automatic COVID-19 detectors should leverage multiple data sources not limited to just images, including patient vitals, population density, geographical location, etc. Image data by itself is typically not sufficient for these types of applications.
For these reasons, I must once again stress that this tutorial is meant for educational purposes only — it is not meant to be a robust COVID-19 detector.
If you believe that yourself or a loved one has COVID-19, you should follow the protocols outlined by the Center for Disease Control (CDC), World Health Organization (WHO), or local country, state, or jurisdiction.
I hope you enjoyed this tutorial and found it educational. It’s also my hope that this tutorial serves as a starting point for anyone interested in applying computer vision and deep learning to automatic COVID-19 detection.
What’s next?
I typically end my blog posts by recommending one of my books/courses, so that you can learn more about applying Computer Vision and Deep Learning to your own projects. Out of respect for the severity of the coronavirus, I am not going to do that — this isn’t the time or the place.
Instead, what I will say is we’re in a very scary season of life right now.
Like all seasons, it will pass, but we need to hunker down and prepare for a cold winter — it’s likely that the worst has yet to come.
To be frank, I feel incredibly depressed and isolated. I see:
- Stock markets tanking.
- Countries locking down their borders.
- Massive sporting events being cancelled.
- Some of the world’s most popular bands postponing their tours.
- And locally, my favorite restaurants and coffee shops shuttering their doors.
That’s all on the macro-level — but what about the micro-level?
What about us as individuals?
It’s too easy to get caught up in the global statistics.
We see numbers like 6,000 dead and 160,000 confirmed cases (with potentially multiple orders of magnitude more due to lack of COVID-19 testing kits and that some people are choosing to self-quarantine).
When we think in those terms we lose sight of ourselves and our loved ones. We need to take things day-by-day. We need to think at the individual level for our own mental health and sanity. We need safe spaces where we can retreat to.
When I started PyImageSearch over 5 years ago, I knew it was going to be a safe space. I set the example for what PyImageSearch was to become and I still do to this day. For this reason, I don’t allow harassment in any shape or form, including, but not limited to, racism, sexism, xenophobia, elitism, bullying, etc.
The PyImageSearch community is special. People here respect others — and if they don’t, I remove them.
Perhaps one of my favorite displays of kind, accepting, and altruistic human character came when I ran PyImageConf 2018 — attendees were overwhelmed with how friendly and welcoming the conference was.
Dave Snowdon, software engineer and PyImageConf attendee said:
PyImageConf was without a doubt the most friendly and welcoming conference I’ve been to. The technical content was also great too! It was privilege to meet and learn from some of the people who’ve contributed their time to build the tools that we rely on for our work (and play).
David Stone, Doctor of Engineering and professor at Virginia Commonwealth University shared the following:
Thanks for putting together PyImageConf. I also agree that it was the most friendly conference that I have attended.
Why do I say all this?
Because I know you may be scared right now.
I know you might be at your whits end (trust me, I am too).
And most importantly, because I want PyImageSearch to be your safe space.
- You might be a student home from school after your semester prematurely ended, disappointed that your education has been put on hold.
- You may be a developer, totally lost after your workplace chained its doors for the foreseeable future.
- You may be a researcher, frustrated that you can’t continue your experiments and authoring that novel paper.
- You might be a parent, trying, unsuccessfully, to juggle two kids and a mandatory “work from home” requirement.
Or, you may be like me — just trying to get through the day by learning a new skill, algorithm, or technique.
I’ve received a number of emails from PyImageSearch readers who want to use this downtime to study Computer Vision and Deep Learning rather than going stir crazy in their homes.
I respect that and I want to help, and to a degree, I believe it is my moral obligation to help how I can:
- To start, there are over 350+ free tutorials you can learn from on the PyImageSearch blog. I publish a new tutorial every Monday at 10AM EST.
- I’ve categorized, cross-referenced, and compiled these tutorials on my “Get Started” page.
- The most popular topics on the “Get Started” page include “Deep Learning” and “Face Applications”.
All these guides are 100% free. Use them to study and learn from.
That said, many readers have also been requesting that I run a sale on my books and courses. At first, I was a bit hesitant about it — the last thing I want is for people to think I’m somehow using the coronavirus as a scheme to “make money”.
But the truth is, being a small business owner who is not only responsible for myself and my family, but the lives and families of my teammates, can be terrifying and overwhelming at times — peoples lives, including small businesses, will be destroyed by this virus.
To that end, just like:
- Bands and performers are offering discounted “online only” shows
- Restaurants are offering home delivery
- Fitness coaches are offering training sessions online
…I’ll be following suit.
Starting tomorrow I’ll be running a sale on PyImageSearch books. This sale isn’t meant for profit and it’s certainly not planned (I’ve spent my entire weekend, sick, trying to put all this together).
Instead, it’s sale to help people, like me (and perhaps like yourself), who are struggling to find their safe space during this mess. Let myself and PyImageSearch become your retreat.
I typically only run one big sale per year (Black Friday), but given how many people are requesting it, I believe it’s something that I need to do for those who want to use this downtime to study and/or as a distraction from the rest of the world.
Feel free to join in or not. It’s totally okay. We all process these tough times in our own ways.
But if you need rest, if you need a haven, if you need a retreat through education — I’ll be here.
Thank you and stay safe.
What's next? We recommend PyImageSearch University.
86+ total classes • 115+ hours hours of on-demand code walkthrough videos • Last updated: February 2026
★★★★★ 4.84 (128 Ratings) • 16,000+ Students Enrolled
I strongly believe that if you had the right teacher you could master computer vision and deep learning.
Do you think learning computer vision and deep learning has to be time-consuming, overwhelming, and complicated? Or has to involve complex mathematics and equations? Or requires a degree in computer science?
That’s not the case.
All you need to master computer vision and deep learning is for someone to explain things to you in simple, intuitive terms. And that’s exactly what I do. My mission is to change education and how complex Artificial Intelligence topics are taught.
If you're serious about learning computer vision, your next stop should be PyImageSearch University, the most comprehensive computer vision, deep learning, and OpenCV course online today. Here you’ll learn how to successfully and confidently apply computer vision to your work, research, and projects. Join me in computer vision mastery.
Inside PyImageSearch University you'll find:
- ✓ 86+ courses on essential computer vision, deep learning, and OpenCV topics
- ✓ 86 Certificates of Completion
- ✓ 115+ hours hours of on-demand video
- ✓ Brand new courses released regularly, ensuring you can keep up with state-of-the-art techniques
- ✓ Pre-configured Jupyter Notebooks in Google Colab
- ✓ Run all code examples in your web browser — works on Windows, macOS, and Linux (no dev environment configuration required!)
- ✓ Access to centralized code repos for all 540+ tutorials on PyImageSearch
- ✓ Easy one-click downloads for code, datasets, pre-trained models, etc.
- ✓ Access on mobile, laptop, desktop, etc.
Summary
In this tutorial you learned how you could use Keras, TensorFlow, and Deep Learning to train an automatic COVID-19 detector on a dataset of X-ray images.
High quality, peer reviewed image datasets for COVID-19 don’t exist (yet), so we had to work with what we had, namely Joseph Cohen’s GitHub repo of open-source X-ray images:
- We sampled 25 images from Cohen’s dataset, taking only the posterioranterior (PA) view of COVID-19 positive cases.
- We then sampled 25 images of healthy patients using Kaggle’s Chest X-Ray Images (Pneumonia) dataset.
From there we used Keras and TensorFlow to train a COVID-19 detector that was capable of obtaining 90-92% accuracy on our testing set with 100% sensitivity and 80% specificity (given our limited dataset).
Keep in mind that the COVID-19 detector covered in this tutorial is for educational purposes only (refer to my “Disclaimer” at the top of this tutorial). My goal is to inspire deep learning practitioners, such as yourself, and open your eyes to how deep learning and computer vision can make a big impact on the world.
I hope you enjoyed this blog post.
To download the source code to this post (including the pre-trained COVID-19 diagnosis model), just enter your email address in the form below!
Download the Source Code and FREE 17-page Resource Guide
Enter your email address below to get a .zip of the code and a FREE 17-page Resource Guide on Computer Vision, OpenCV, and Deep Learning. Inside you'll find my hand-picked tutorials, books, courses, and libraries to help you master CV and DL!

